Understanding how a drug interacts with the protein it is designed to target is a critical step in drug discovery. A drug must hit the right target with the desired effect before it can be considered safe to enter clinical trials.
Structure-based drug design (SBDD) is Our company's approach to the design and optimization of chemical structures. Our goal is to use the knowledge of a candidate's three-dimensional (3D) structure and how its shape and charge allow it to interact with its biological target to determine the most suitable compound for clinical testing.
Service Overview
Our company has knowledge of the 3D structure of a biological target obtained by methods such as X-ray crystallography or NMR spectroscopy. If the experimental structure of the target is not available, we can create a homology model of the target based on the experimental structure of the protein in question.
We use the structure of the biological target, and interactive graphics and intuitive designs for medical chemists can be used to predict drug candidates with high affinity and selective binding to the target. Various automated computational programs can be used to suggest novel drug candidates.
Services Contents
- High throughput screening and active compound discovery based on structure design.
- Discovery of active compounds to lead compounds.
- Optimization of lead compounds to identify preclinical drug candidates.
- Structure-activity relationship studies.
Research Capabilities
Our company's current structure-based drug design approaches fall into three broad categories.
Method 1
We search for new receptor ligands in a large database of small molecule 3D structures in order to find suitable ligands for receptor binding pockets using a rapid proximity docking procedure. This approach is known as virtual screening.
Method 2: Ab initio design of new ligands
We build ligand molecules by stepwise assembly of small fragments within the constraints of the binding pocket. These fragments can be individual atoms or molecular fragments. The main advantage of this approach is that it allows us to propose new structures that are not found in any database.
Method 3
We optimize known ligands by evaluating the analogs proposed in the binding cavity.
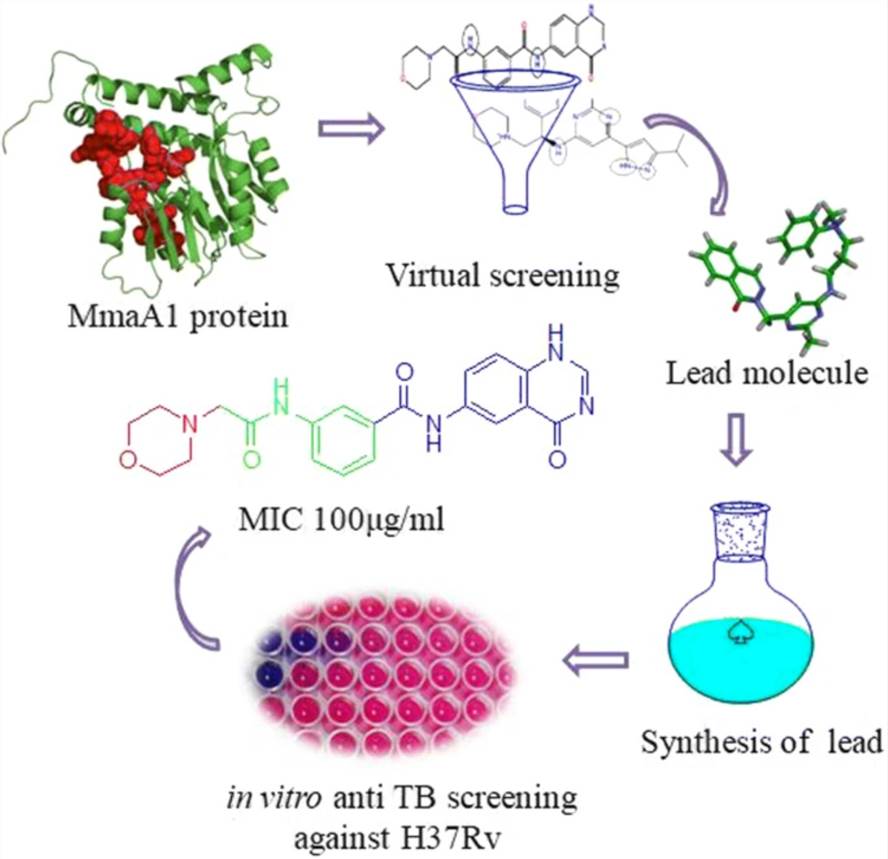 Fig.1 Structure based virtual screening study was performed using a validated homology model against small molecules from two virtual compound libraries. (Veeravarapu H, et al. 2021)
Fig.1 Structure based virtual screening study was performed using a validated homology model against small molecules from two virtual compound libraries. (Veeravarapu H, et al. 2021)
Sample Requirements
(1) The sample needs to have good steric and electronic complementarity with the target biomolecule.
(2) To bind tightly, a significant amount of hydrophobic surface should be buried in the complex.
(3) Sufficient conformational rigidity is essential to ensure that the entropy loss upon ligand binding is acceptable.
(4) There are at least three additional criteria to consider in the drug design cycle.
(5) The sample is chemically stable.
(6) Sufficient solubility in water for inhibition tests and structural studies.
(7) Ease of synthesis, including avoidance of chiral centers and "dead-end leads".
Our Advantages
Cryo-electron Microscopic Structure Analysis
We are a world leader in the research and application of cryo-electron microscopic structure resolution techniques. Cryo-electron microscopy (Cryo-EM) can provide the necessary insight into
- The biological function of proteins.
- The role of proteins in disease.
- How drug candidates bind and interact with proteins.
Rich Database Resources
We have a library of over 600 million drug-forming molecules, which makes it much easier to locate active chemicals while performing computational chemistry screening.
Efficient and Flexible Computational Platform
We may flexibly select the service mode of cloud computing and high-performance computing, and we can dynamically extend our computing resources in accordance with the various computing requirements of customers.
If you are looking for smarter, higher quality solutions that incorporate best practices, please feel free to contact us.
Reference
- Veeravarapu H, et al. (2021). "Structure-based Drug Design, Synthesis and Screening of MmaA1 Inhibitors as Novel Anti-TB Agents." Molecular Diversity. 25: 351-366.
Related Services
It should be noted that our service is only used for research, not for clinical use.

 Fig.1 Structure based virtual screening study was performed using a validated homology model against small molecules from two virtual compound libraries. (Veeravarapu H, et al. 2021)
Fig.1 Structure based virtual screening study was performed using a validated homology model against small molecules from two virtual compound libraries. (Veeravarapu H, et al. 2021)